概要
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet 1 | RosetteSep™ DM-L Density Medium | 15705 | All | MULTI |
| Product Information Sheet 2 | RosetteSep™ DM-L Density Medium | 15705 | All | English |
| Safety Data Sheet | RosetteSep™ DM-L Density Medium | 15705 | All | English |
数据及文献
Data
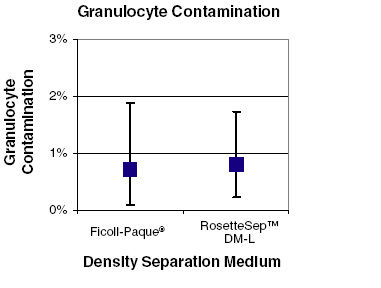
Figure 1. Minimal Granulocyte Contamination Using RosetteSep™ DM-L Density Medium
Granulocyte contamination is typically less than 1% when enriching lymphocytes from fresh whole blood using RosetteSep™. There was no significant difference in the level of granulocyte contamination when using RosetteSep™ DM-L in place of Ficoll-paque PLUS to enrich lymphocytes (N=11)
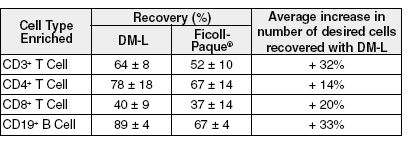
Figure 2. Rosettesep™ DM-L Improves the Recovery of Lymphocytes Enriched with Rosettesep™ from Fresh Blood
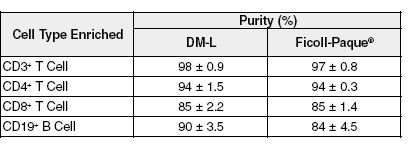
Figure 3. Cell Purity Is Equivalent or Higher for Rosettesep™ Lymphocyte Enrichment Using Rosettesep™ DM-L Instead of Ficoll-paque®

 网站首页
网站首页