概要
SepMate™ is a tube that facilitates the isolation of PBMCs or specific cell types by density gradient centrifugation. SepMate™ tubes contain an insert that provides a barrier between the density gradient medium and blood. SepMate™ eliminates the need for careful layering of blood onto the density gradient medium, and allows for fast and easy harvesting of the isolated mononuclear cells with a simple pour. SepMate™ is also compatible with RosetteSep™ enrichment cocktails, allowing isolation of specific cell types in less than 30 minutes.
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | SepMate™-15 (IVD) | 85415, 85420 | All | English |
数据及文献
Data
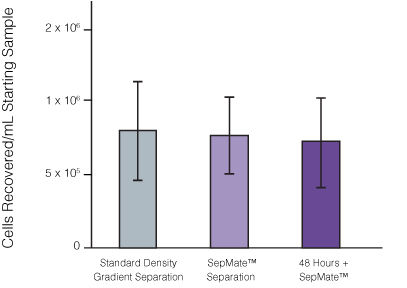
Figure 1. Recovery of mononuclear cells (MNCs) from peripheral blood using SepMate™-50 versus standard density gradient centrifiguation.
Recovery of MNCs from fresh and 48-hour post blood draw enriched by density gradient centrifugation with SepMate™ (purple) or without (grey). There was no significant difference in the recovery of MNCS with and without SepMate™.
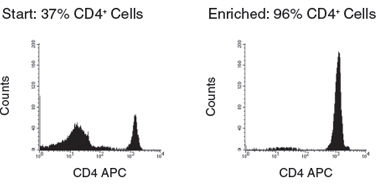
Figure 2. Human CD4+ T Cell Isolation using SepMate™-50 and RosetteSep™ Human CD4+ T Cell Enrichment Cocktail

 网站首页
网站首页