概要
The EasySep™ Release Human PE Positive Selection Kit is designed to isolate cells that are labeled with PE-conjugated antibodies from fresh or previously frozen peripheral blood mononuclear cells or washed leukapheresis samples by immunomagnetic positive selection.
Desired cells are labeled with antibodies and magnetic particles, and separated without columns using an EasySep™ magnet. Unwanted cells are simply poured off, while desired cells remain in the tube. Then, bound magnetic particles are removed from the EasySep™-isolated, PE-antibody labeled cells, which are immediately available for downstream applications. Following cell isolation with this EasySep™ Release kit, antibody complexes remain bound to the cell surface and may interact with Brilliant Violet™ antibody conjugates, polyethylene glycol-modified proteins or other chemically related ligands.
This product replaces the EasySep™ Human PE Positive Selection Kit (Catalog #18551), providing highly purified particle-free cells.
Desired cells are labeled with antibodies and magnetic particles, and separated without columns using an EasySep™ magnet. Unwanted cells are simply poured off, while desired cells remain in the tube. Then, bound magnetic particles are removed from the EasySep™-isolated, PE-antibody labeled cells, which are immediately available for downstream applications. Following cell isolation with this EasySep™ Release kit, antibody complexes remain bound to the cell surface and may interact with Brilliant Violet™ antibody conjugates, polyethylene glycol-modified proteins or other chemically related ligands.
This product replaces the EasySep™ Human PE Positive Selection Kit (Catalog #18551), providing highly purified particle-free cells.
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet 1 | EasySep™ Release Human PE Positive Selection Kit | 17654 | 19E102695 or lower | English |
| Product Information Sheet 2 | EasySep™ Release Human PE Positive Selection Kit | 17654 | 1000005896 and higher | English |
| Safety Data Sheet 1 | EasySep™ Release Human PE Positive Selection Kit | 17654 | All | English |
| Safety Data Sheet 2 | EasySep™ Release Human PE Positive Selection Kit | 17654 | All | English |
| Safety Data Sheet 3 | EasySep™ Release Human PE Positive Selection Kit | 17654 | All | English |
| Safety Data Sheet 4 | EasySep™ Release Human PE Positive Selection Kit | 17654 | All | English |
数据及文献
Data
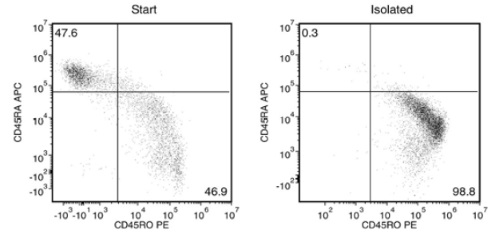
Starting with fresh PBMCs, the purities of the start and final isolated fractions are 46.9% and 98.8%, respectively, using a PE-conjugated anti-human CD45RO antibody and EasySep™ Release Human Positive Selection Kit.
Publications (2)
Science translational medicine 2020 jul
STING differentially regulates experimental GVHD mediated by CD8 versus CD4 T cell subsets.
Abstract
Abstract
The stimulator of interferon genes (STING) pathway has been proposed as a key regulator of gastrointestinal homeostasis and inflammatory responses. Although STING reportedly protects against gut barrier damage and graft-versus-host disease (GVHD) after major histocompatibility complex (MHC)-mismatched allogeneic hematopoietic stem cell transplantation (aHSCT), its effect in clinically relevant MHC-matched aHSCT is unknown. Studies here demonstrate that STING signaling in nonhematopoietic cells promoted MHC-matched aHSCT-induced GVHD and that STING agonists increased type I interferon and MHC I expression in nonhematopoietic mouse intestinal organoid cultures. Moreover, mice expressing a human STING allele containing three single-nucleotide polymorphisms associated with decreased STING activity also developed reduced MHC-matched GVHD, demonstrating STING's potential clinical importance. STING-/- recipients experienced reduced GVHD with transplant of purified donor CD8+ T cells in both MHC-matched and MHC-mismatched models, reconciling the seemingly disparate results. Further examination revealed that STING deficiency reduced the activation of donor CD8+ T cells early after transplant and promoted recipient MHC class II+ antigen-presenting cell (APC) survival. Therefore, APC persistence in STING pathway absence may account for the increased GVHD mediated by CD4+ T cells in completely mismatched recipients. In total, our findings have important implications for regulating clinical GVHD by targeting STING early after aHSCT and demonstrate that an innate immune pathway has opposing effects on the outcome of aHSCT, depending on the donor/recipient MHC disparity.
Frontiers in immunology 2020
Rheumatoid Arthritis Patients, Both Newly Diagnosed and Methotrexate Treated, Show More DNA Methylation Differences in CD4+ Memory Than in CD4+ Na\ive T Cells."
Abstract
Abstract
Background: Differences in DNA methylation have been reported in B and T lymphocyte populations, including CD4+ T cells, isolated from rheumatoid arthritis (RA) patients when compared to healthy controls. CD4+ T cells are a heterogeneous cell type with subpopulations displaying distinct DNA methylation patterns. In this study, we investigated DNA methylation using reduced representation bisulfite sequencing in two CD4+ T cell populations (CD4+ memory and na{\{i}}ve cells) in three groups: newly diagnosed disease modifying antirheumatic drugs (DMARD) na{\"{i}}ve RA patients (N = 11) methotrexate (MTX) treated RA patients (N = 18) and healthy controls (N = 9) matched for age gender and smoking status. Results: Analyses of these data revealed significantly more differentially methylated positions (DMPs) in CD4+ memory than in CD4+ na{\""{i}}ve T cells (904 vs. 19 DMPs) in RA patients compared to controls. The majority of DMPs (72{\%}) identified in newly diagnosed and DMARD na{\""{i}}ve RA patients with active disease showed increased DNA methylation (39 DMPs) whereas most DMPs (80{\%}) identified in the MTX treated RA patients in remission displayed decreased DNA methylation (694 DMPs). Interestingly we also found that about one third of the 101 known RA risk loci overlapped (±500 kb) with the DMPs. Notably introns of the UBASH3A gene harbor both the lead RA risk SNP and two DMPs in CD4+ memory T cells. Conclusion: Our results suggest that RA associated DNA methylation differences vary between the two T cell subsets but are also influenced by RA characteristics such as disease activity disease duration and/or MTX treatment."""

 网站首页
网站首页



