概要
The EasySep™ Mouse CD19 Positive Selection Kit II is designed to isolate CD19+ cells from single-cell suspensions of splenocytes or other tissues by positive selection. Desired cells are targeted with antibody complexes recognizing CD19 and dextran-coated magnetic particles. Labeled cells are separated using an EasySep™ magnet without the use of columns. Cells of interest remain in the tube while unwanted cells are poured off.
This product replaces the EasySep™ Mouse CD19 Positive Selection Kit (Catalog #18754) for even faster cell isolations and does not result in the labeling of isolated cells with PE.
This product replaces the EasySep™ Mouse CD19 Positive Selection Kit (Catalog #18754) for even faster cell isolations and does not result in the labeling of isolated cells with PE.
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | EasySep™ Mouse CD19 Positive Selection Kit II | 18954 | All | English |
| Product Information Sheet | RoboSep™ Mouse CD19 Positive Selection Kit II | 18954RF | All | English |
| Safety Data Sheet 1 | EasySep™ Mouse CD19 Positive Selection Kit II | 18954 | All | English |
| Safety Data Sheet 2 | EasySep™ Mouse CD19 Positive Selection Kit II | 18954 | All | English |
| Safety Data Sheet 3 | EasySep™ Mouse CD19 Positive Selection Kit II | 18954 | All | English |
数据及文献
Data
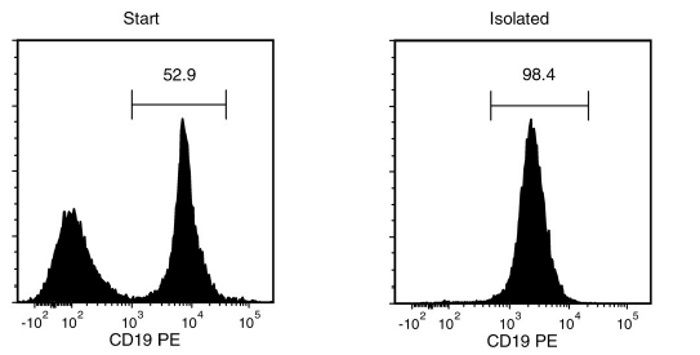
Figure 1. Typical EasySep™ CD19 Positive Selection Profile
Starting with mouse splenocytes, the CD19+ cell content of the isolated fraction is typically 98.1 ± 0.6% (mean ± SD using the purple EasySep™ Magnet).
Publications (2)
Cell reports 2019 dec
Discovery of Small Molecules for the Reversal of T Cell Exhaustion.
Abstract
Abstract
Inhibitory receptors (IRs) function as critical regulators of immune responses by tempering T cell activity. In humans, several persisting viruses as well as cancers exploit IR signaling by upregulating IR ligands, resulting in suppression of T cell function (i.e., exhaustion). This allows escape from immune surveillance and continuation of disease. Here, we report the design, implementation, and results of a phenotypic high-throughput screen for molecules that modulate CD8+ T cell activity. We identify 19 compounds from the ReFRAME drug-repurposing collection that restore cytokine production and enhance the proliferation of exhausted T cells. Analysis of our top hit, ingenol mebutate, a protein kinase C (PKC) inducing diterpene ester, reveals a role for this molecule in overriding the suppressive signaling cascade mediated by IR signaling on T cells. Collectively, these results demonstrate a disease-relevant methodology for identifying modulators of T cell function and reveal new targets for immunotherapy.
Genes and immunity 2016 SEP
Natural genetic variation profoundly regulates gene expression in immune cells and dictates susceptibility to CNS autoimmunity.
Abstract
Abstract
Regulation of gene expression in immune cells is known to be under genetic control, and likely contributes to susceptibility to autoimmune diseases such as multiple sclerosis (MS). How this occurs in concert across multiple immune cell types is poorly understood. Using a mouse model that harnesses the genetic diversity of wild-derived mice, more accurately reflecting genetically diverse human populations, we provide an extensive characterization of the genetic regulation of gene expression in five different naive immune cell types relevant to MS. The immune cell transcriptome is shown to be under profound genetic control, exhibiting diverse patterns: global, cell-specific and sex-specific. Bioinformatic analysis of the genetically controlled transcript networks reveals reduced cell type specificity and inflammatory activity in wild-derived PWD/PhJ mice, compared with the conventional laboratory strain C57BL/6J. Additionally, candidate MS-GWAS (genome-wide association study candidate genes for MS susceptibility) genes were significantly enriched among transcripts overrepresented in C57BL/6J cells compared with PWD. These expression level differences correlate with robust differences in susceptibility to experimental autoimmune encephalomyelitis, the principal model of MS, and skewing of the encephalitogenic T-cell responses. Taken together, our results provide functional insights into the genetic regulation of the immune transcriptome, and shed light on how this in turn contributes to susceptibility to autoimmune disease.Genes and Immunity advance online publication, 22 September 2016; doi:10.1038/gene.2016.37.

 网站首页
网站首页