概要
StemSpan™ CD34+ Expansion Supplement typically promotes ~40-fold expansion of total nucleated cells in 7-day liquid cultures of CD34+ human cord blood (CB) cells. After one week, ~40% of the cultured cells express CD34, indicative of >10-fold expansion of input CD34+ CB cells. This expansion may be further increased with the addition of small molecules such as UM729. See data tab for more details.
StemSpan™ CD34+ Expansion Supplement (10X) is intended for use in combination with any of the following StemSpan™ media:
• StemSpan™ SFEM (Catalog #09600)
• StemSpan™ SFEM II (Catalog #09605)
• StemSpan™-XF (Catalog #100-0073)
• StemSpan™-AOF (Catalog #100-0130)
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | StemSpan™ CD34+ Expansion Supplement (10X) | 02691 | All | English |
| Safety Data Sheet | StemSpan™ CD34+ Expansion Supplement (10X) | 02691 | All | English |
数据及文献
Data
Table 1. HSC Expansion Culture with CD34+ Human Cord Blood Cells Cultured in StemSpan™ SFEM Containing CD34+ Expansion Supplement
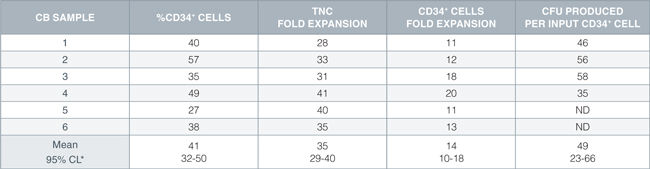
Shown are the percent CD34+ cells, fold expansion of total nucleated cells (TNC) and CD34+ cells, and numbers of colony-forming units (CFU) produced per input CD34+ cell after 7 days of hsc expansion culture of enriched CD34+ cells from six independent cord blood (CB) samples.
*95% confidence limits, the range within which 95% of the results will typically fall.
ND: not done
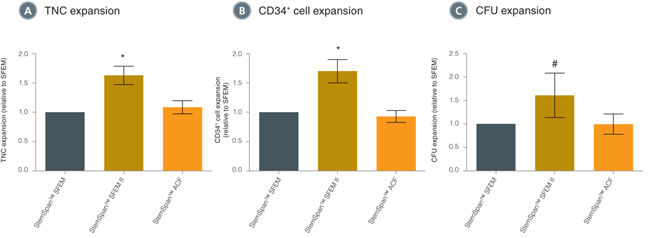
Figure 1. Comparison of CD34+ Cell Expansion in Different StemSpan™ Media Containing CD34+ Expansion Supplement
Average expansion of (A) total nucleated cells (TNC), (B) CD34+ cells and (C) colony-forming units (CFU), normalized relative to the values obtained in StemSpan™ SFEM (grey bars) after culturing purified hematopoietic CD34+ cord blood cells (n=6) for 7 days in StemSpan™ SFEM, SFEM II (gold bars) or ACF (orange bars) media containing CD34+ Expansion Supplement. Vertical lines indicate 95% confidence limits, the range within which 95% of results will typically fall. Cell yields in StemSpan™ SFEM II were on average ~60% higher than in StemSpan™ SFEM and StemSpan™ ACF.
*p<0.001, #p<0.05 (paired t-test, n=6 in A and B, n=4 in C).
Note: Data for StemSpan™-ACF shown were generated with the original phenol red-containing version. However internal testing showed that the performance of the new phenol red-free, cGMP-manufactured version, StemSpan™-AOF (Catalog #100-0130) was comparable.

 网站首页
网站首页