概要
This kit is optimized for differentiation of cells maintained in mTeSR™1. For differentiation of cells maintained in TeSR™-E8™, please see the STEMDiff™ Definitive Endoderm Kit (TeSR™-E8™ Optimized).
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | STEMdiff™ Definitive Endoderm Kit | 05110 | Component #05113: Lot 18D89658 and higher | English |
| Safety Data Sheet 1 | STEMdiff™ Definitive Endoderm Kit | 05110 | All | English |
| Safety Data Sheet 2 | STEMdiff™ Definitive Endoderm Kit | 05110 | All | English |
| Safety Data Sheet 3 | STEMdiff™ Definitive Endoderm Kit | 05110 | All | English |
数据及文献
Data
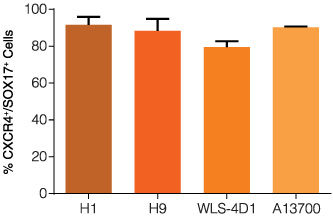
Figure 1. Definitive endoderm differentiation is efficient across multiple human ES and iPS cell lines
Quantitative analysis of definitive endoderm formulation on multiple human ES and iPS cell lines as measured by co-expression of CXCR4 and SOX17. Prior to differentiation using STEMdiff™ Definitive Endoderm, cells were maintained in their pluripotent state by culturing mTeSR™1 on Matrigel. Data are expressed as the mean percent of cells expressing both markers. Error bars indicate SEM, n = 4-18 per cell line.
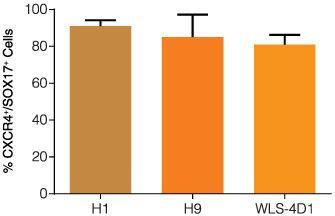
Figure 2. Quantitative Analysis of Definitive Endoderm hES and iPS-Derived Using STEMdiff™ Definitive Endoderm
Quantitative analysis of definitive endoderm in human ES and iPS cells previously maintained in TeSR™2 prior to differentiation on Matrigel using STEMdiff™ Definitive Endoderm. Data are expressed as the mean percent of cells expressing both markers. Error bars indicate SEM. n = 4-11 per cell line.
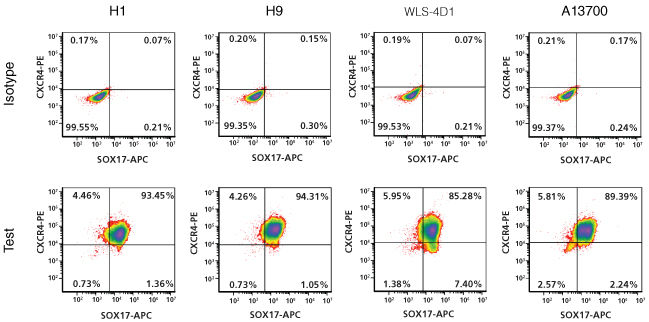
Figure 3. Efficient definitive endoderm differentiation in human ES and iPS cells
Representative Density plots showing CXCR4 and SOX17 expression in human ES cells (H1 and H9) and human iPS cells (WLS-4D1 and A13700) following 5 days of differentiation to definitive endoderm using STEMdiff™ Definitive Endoderm. Isotype controls were used to set quadrant gates.
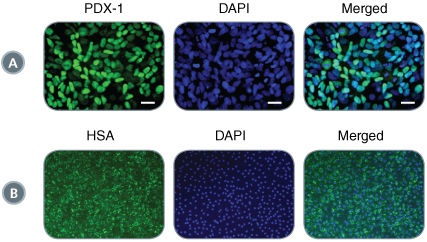
Figure 4. STEMdiff™ Definitive Endoderm yields DE that retains potency for downstream lineage specification
Cultures differentiated using STEMdiff™ Definitive Endoderm maintain their ability to be directed towards pancreatic and hepatic lineages. A) Representative image of PDX-1 immunoreactivity in H9 cells following pancreatic specification. Scale bar 20 µm. B) Representative image of human serum albumin (HSA) immunoreactivity in H9 cells following hepatic specification. Scale bar, 100 µm.
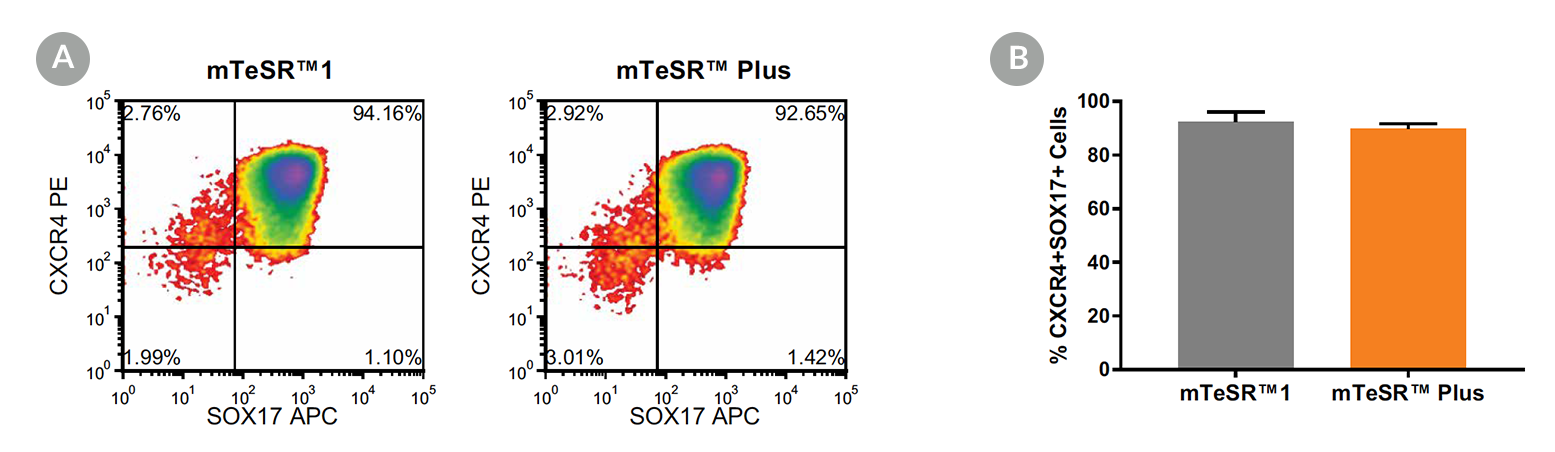
Figure 5. Generation of Definitive Endoderm from hPSCs Maintained in mTeSR™ Plus
(A) Representative density plots showing CXCR4 and SOX17 expression in cells cultured in mTeSR™1 (daily feeds) or mTeSR™ Plus (restricted feeds), following 5 days of differentiation using the STEMdiff™ Definitive Endoderm Kit. (B) Quantitative analysis of definitive endoderm formation in multiple hPSC lines (H9, STiPS-M001, WLS-1C) maintained with mTeSR™1 or mTeSR™ Plus as measured by co-expression of CXCR4 and SOX17. Data are expressed as the mean percentage of cells (± SEM) expressing both markers; n=3.

 网站首页
网站首页